PDFgui¶
Overview¶
For users who do not have the expertise or necessity for command line analysis, PDFgui is a convenient and easy to use graphical front end for the PDFfit2 refinement program. It is capable of full-profile fitting of the atomic pair distribution function (PDF) derived from x-ray or neutron diffraction data and comes with built in graphical and structure visualization capabilities.
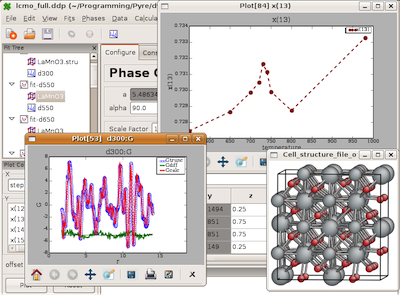
Screen-shot of PDFgui during a refinement of multiple data-sets¶
Reference¶
If you use this program for a scientific research that leads to publication, we ask that you acknowledge use of the program by citing the following paper in your publication:
C L Farrow, P Juhas, J W Liu, D Bryndin, E S Božin, J Bloch, Th Proffen and S J L Billinge, PDFfit2 and PDFgui: computer programs for studying nanostructure in crystals, J. Phys.: Condens. Matter 19 (2007) 335219.
Installation¶
By downloading and using this software you are agreeing to the conditions specified in the software license.
PDFgui is distributed as a software package for Anaconda Python. PDFgui is available for all operating systems supported by Anaconda, namely for Linux, Mac OS X, and Windows.
As a prerequisite for PDFgui installation, download and install Miniconda (https://docs.conda.io/en/latest/miniconda.html#latest-miniconda-installer-links) or Anaconda Python (https://www.anaconda.com/download).
PDFgui is available from the “conda-forge” channel of Anaconda packages.
There are currently two different sets of instructions to install it depending on your platform. If you want to install it on Windows, Linux, or a non-arm64 Mac OSX (i.e., if at the time of writing your Mac does not have an M1 or M2 chip) use the first set, otherwise the second set of instructions
Windows, macOS (non-Arm64), Linux¶
Add the “conda-forge” channel by running the following command in a terminal:
conda config --add channels conda-forge
Create a new environment named diffpy.pdfgui_env (or any name of your choice) and
install diffpy.pdfgui:
conda create -n diffpy.pdfgui_env diffpy.pdfgui
Activate the environment:
conda activate diffpy.pdfgui_env
Confirm that the installation was successful:
python -c "import diffpy.pdfgui; print(diffpy.pdfgui.__version__)"
macOS (Arm64)¶
Create a new conda environment diffpy.pdfgui_env:
conda config --add channels conda-forge
conda create -n diffpy.pdfgui_env python=3.13
Activate the environment:
conda activate diffpy.pdfgui_env
It is necessary to get versions of pdffit2 built for Mac from Python package index (Pypi). First we will install all the dependencies from conda-forge and then the pdffit2 itself from Pypi using the following commands:
conda install wxpython diffpy.utils matplotlib-base pycifrw
pip install diffpy.pdffit2
Finally, we want to install PDFgui, again from conda-forge:
pip install diffpy.pdfgui
Running pdfgui:¶
Note
To start PDFgui from a Terminal make sure pdfgui_env is the active Anaconda environment:
conda activate pdfgui_env
Alternatively, on bash terminals you can add the following line to your shell startup
file .bashrc to define an alias which will work in
any Anaconda environment
alias pdfgui="/path/to/pdfgui_env/bin/pdfgui"
where /path/to needs to be adjusted according to the output of
which pdfgui command.
Please consider joining the diffpy-users Google group. News about updates and new releases will be made there, as well as it being a place to ask questions and find answers about using all diffpy programs. When you sign up, please leave a short message about why you are requesting to join as we have had spam accounts requesting access and so we want to know you are a legitimate user.
Documentation and help¶
Search “PDFgui” on YouTube for some video tutorials on how to use PDFgui.
For in-depth help in using PDFgui to solve scientific problems please see the book “Atomic Pair Distribution Function Analysis: A primer” by Simon Billinge, Kirsten Jensen, and past and present Billinge group members, published by Oxford University Press. Data for the worked examples can be found here: https://github.com/Billingegroup/pdfttp_data
Tutorial files are available from pdfgui-tutorial.zip.
Please, join the community forum for tips, tricks, and feedback.
Legacy versions¶
v1.1.2 is the last python 2 version of PDFgui. It is no longer supported, but may be needed to open python 2 generated .ddp project files. It can be installed in a python2 anaconda environment using the commands:
conda config --set restore_free_channel true
conda config --add channels diffpy
conda create --name=py27 python=2.7
conda activate py27
conda install -c diffpy "diffpy.pdfgui==1.1.2"
When Anaconda Python is not available, PDFgui can be installed from sources. The latest source package is at the Python Package Index and the prior versions are at https://github.com/diffpy/diffpy.pdfgui/releases. See the README document for further details on installation from sources,
Finally, here are the previous single-file installers. Although quite outdated, they might be handy if there is some problem with Anaconda or if one needs to install without Internet connection.
Name |
Date |
Size |
Description |
|---|---|---|---|
2009-04-10 |
21.5 M |
Windows self extracting installer |
|
2009-04-10 |
6.4 M |
Linux and Mac tarball |
MD5 check sums for these installer files are available here.